welcome to spirochase
This website is an easy-to-navigate
portal to all reference proteomes within the Spirochaetes phylum.
Here, proteins are organized into orthologous groups (OGs), each
with a unique identifier and annotation.
With Spirochase, you can easily find answers to questions like:
Is my protein found in spirochetes?
Is it ancestral to the entire phylum?
How is it distributed across the phylogeny?
You can also explore the data directly on the
Data page
or dive into the latest
phylogeny of Spirochaetota on the
Tree page.
Learn more about what an OG is in
omabrowser
For details on how the data was generated, refer to the study by Huete SG et al. 2025,
available
here (PrePrint)
.
database in numbers
team
Biology of Spirochetes Unit, Institut Pasteur website link

Samuel García Huete, PhD

Mathieu Picardeau, PhD

Nadia Benaroudj, PhD
run analysis
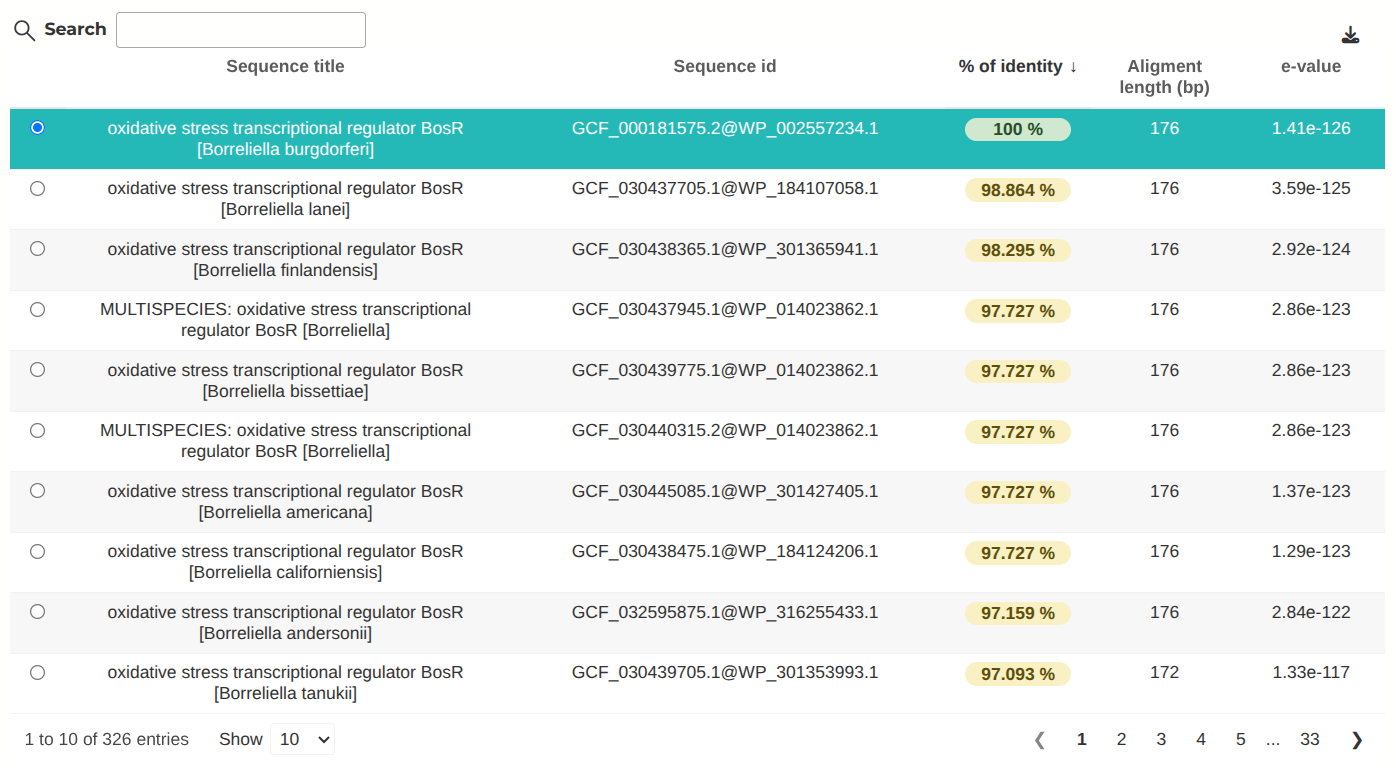
The first step to find out the distribution of your protein of interest is to identify the OG group to which it belongs. We can do this by searching for the protein against the Spirochaetes database.
Here, you can perform a BLAST search for your protein of interest using two databases:
- The SpiroREF database includes all proteins from the reference proteomes of Spirochaetes , ensuring you find all matching hits, though it may return many results from different orthologous groups (OGs). Usually, the first result would be the best match, but you will need to check the statistics presented after the search (e-value, for example)
- The SpiroREP database, on the other hand, contains just one representative sequence per OG, helping you identify the OG your sequence belongs to, but it may not yield high identity matches. In this case, you will usually see a great difference between the best match and all the other hits by checking the statistics presented after the search (e-value, for example).
If in doubt of which database to use, we advise you to use SpiroREP for a first search and, only if you find very similar statistics between two different OGs, you can confirm by searching with SpiroREF. Please note that for a protein to be part of an OG, it must be present in at least two reference proteomes, so SpiroREP may not return significant hits for poorly conserved proteins. To learn more about how the reference genomes were chosen, see the study by Huete SG et al. 2025 . To learn more about how BLAST works, check here .
See the description above
OR
tree page
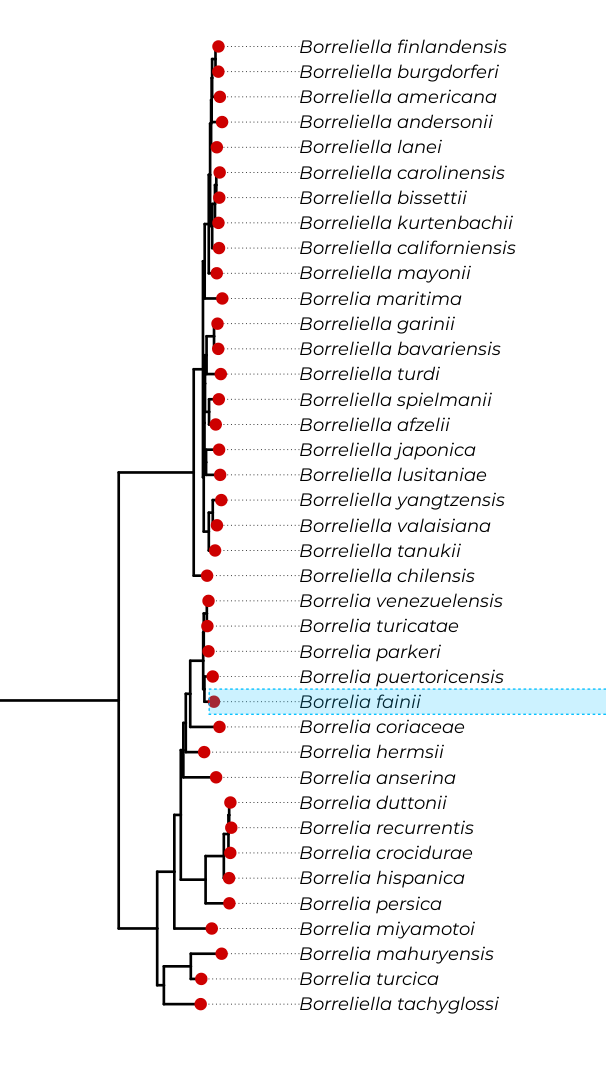
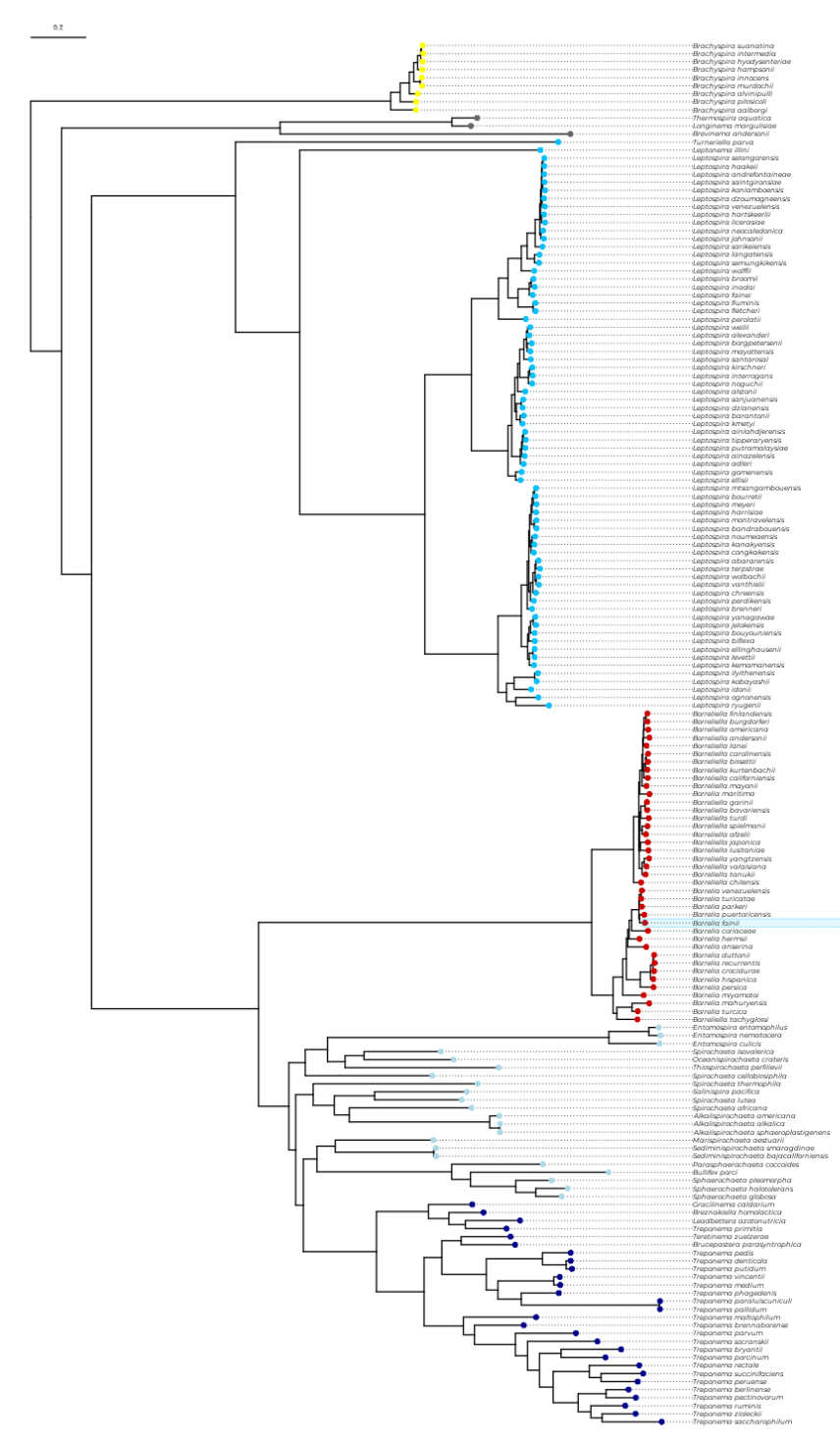
Here, you can explore the latest updated phylogeny of the Spirochaetes phylum, as well as plot the distribution of your OG of interest. From its distribution, you can perhaps infer its function in a process called phylogenetic profiling. To learn more about this phylogeny and how phylogenetic profiling can be applied to identify the function of uncharacterized genes in Spirochaetes , check the study by Huete SG et al. 2025 .
Data Exploration
Here, you can explore the latest annotations of the reference proteomes in Spirochaetes . The interactive graph below displays the percentage of the genome of each species that is dedicated to a given COG category. By selecting or unselecting different genera, you can easily compare their relative COG distribution. See for the download of the complete annotation dataset.
Contacts
TEAM
Samuel García Huete
samuel.garcia-huete.ext@pasteur.fr
PHD
Biology of Spirochetes,
CNRS UMR6047
Institut Pasteur, Paris
Mathieu Picardeau
Research director
Biology of Spirochetes,
CNRS UMR6047
Institut Pasteur, Paris
PUBLICATION
submisson publication in progress
S G. Huete, K Coullin, E Chapeaublanc, R Torchet, N Benaroudj, M Picardeau, Linking genomic evolutionary transitions to ecological phenotypic adaptations in Spirochaetes PrePrint bioRxiv https://doi.org/10.1101/2025.07.04.663154
TEAM WEBSITE
CREDITS
CONTRIBUTORS
Samuel García Huete
samuel.garcia-huete@pasteur.fr
Scientific code
PHD
Biology of Spirochetes,
CNRS UMR6047,
Institut Pasteur, Paris
Rachel Torchet
User Interface and User Experience
UX/UI Designer
Scientific Information Systems and Technical Expertise
Bioinformatics and Biostatistics HUB,
Institut Pasteur, Paris
Elodie Chapeaublanc
elodie.chapeaublanc@pasteur.fr
Implementation and Deployment
Research Engineer - Web Developer
Head of WISPR pole
Bioinformatics and Biostatistics HUB,
Institut Pasteur, Paris
FUNDING
This work was supported by
Error 404
The requested URL was not found - Please go back to home page part